Epigenetic control of cell states and cell identity
Our lab is focused on investigating how cell identity is established, maintained and disrupted. We want to understand how the epigenome controls cell fate. During the development of a multicellular organism a single cell can give rise to an entire organism made of a multitude of cells of specialized function. Such functional specification is determined both by the levels of expression of transcription factors and the receptivity of the chromatin state of the target genes to the binding of such factors. Once cell identity is established, epigenetic modifications, including those on histones and DNA, can ensure that it is stably maintained through cell divisions.
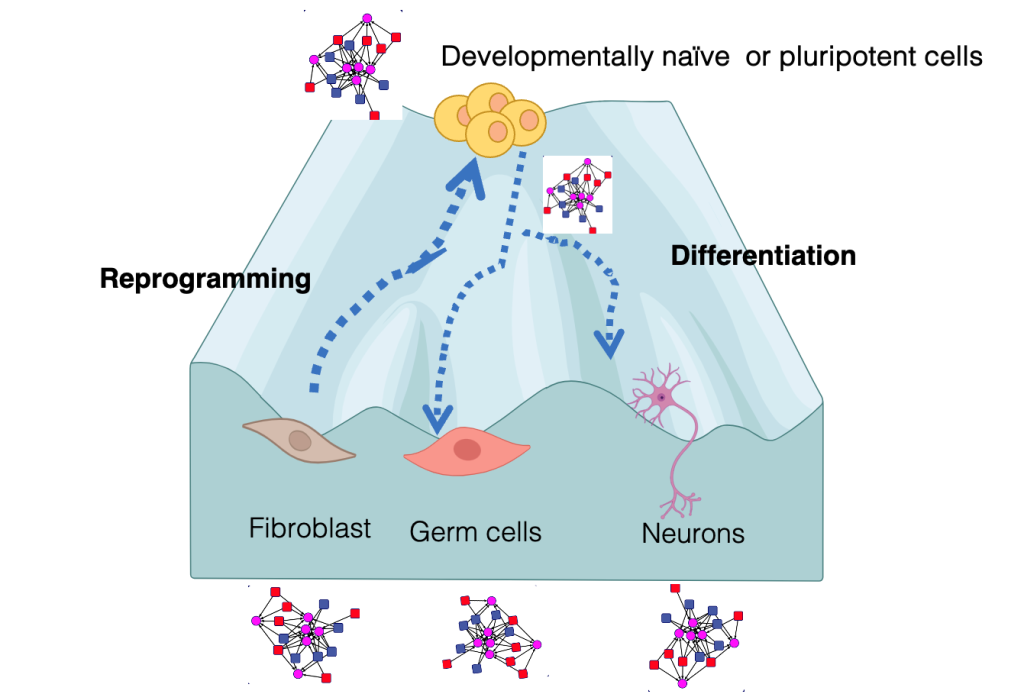
Paradigms to model cell identity
While during development, spatial and morphogenetic cues are integrated to coax cells down a particular lineage, cell fate can be altered in vitro by the overexpression of proteins from other cell types. One of the most dramatic examples of such cell fate conversion is the reprogramming of unipotent somatic cells to induced pluripotent stem cells (iPSCs). iPSCs can self-renew indefinitely and differentiate into all other cell types like embryonic stem cells but bypass the ethical and practical constraints of using ESCs for regenerative therapy. An additional property of iPSCs is the rejuvenation of old somatic cells to a more youthful cell state. We investigate whether the cell states of pluripotency, and rejuvenation can be unentangled. iPSCs share their agelessness with germ cells. We want to understand if special chromatin structures safeguard primordial germ cells from passing on epigenetic memory to the next generation. Primordial germ cells surprisingly share pathways with neuronal differentiation. We will define whether shared epigenetic regulators control the decision points of germ cell versus neuronal cell identity.
Techniques and Methods
We use pharmacological and gene-editing perturbations to confer desired epigenetic properties genome wide or in a site-specific manner (CRISPR-I). We use genome wide techniques to query the epigenome (ChIP-seq, ATAC-seq), transcriptome (RNA-seq) and proteome (IP and histone mass spectrometry) at the population and single cell level (sc RNA, snATAC, scMultiome). With these data we apply computational tools to molecularly define cell states, points of no return, and efficient methods to generate desired cell identity.
 Feedback, questions or accessibility issues:
Feedback, questions or accessibility issues: